History of Nomenclatural Background
Taxonomy of the Genus Laurencia
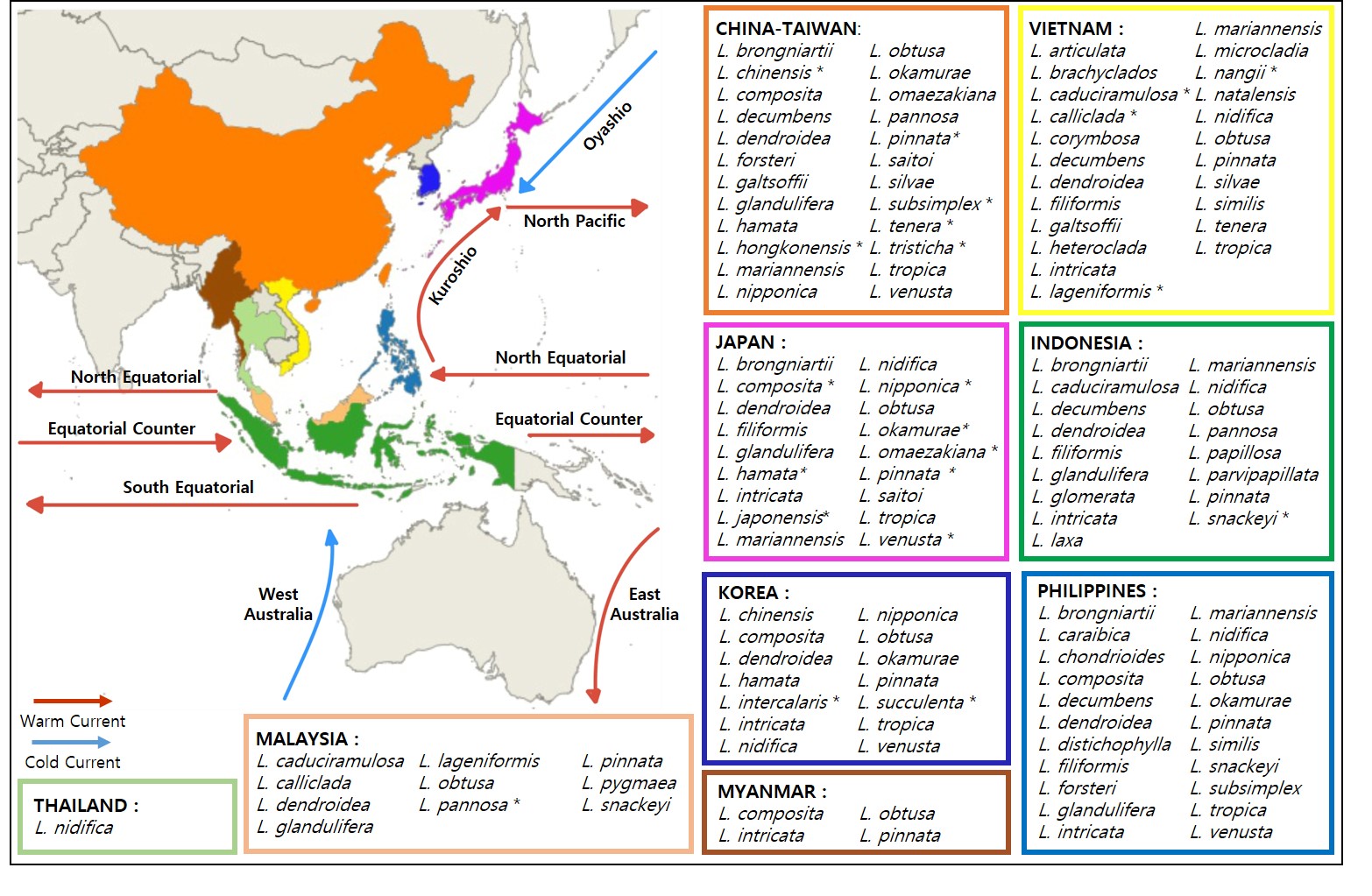
Diversity and Distribution in East Asia and Southeast Asia regions
Molecular Phylogenetic Research
Life History
Ecological Role and Utilization
Declaration of Competing Interest
History of Nomenclatural Background
The red algal genus Laurencia J.V. Lamoroux belongs to the family Rhodomelaceae (Ceramiales, Rhodophyta). The term “Laurencia” was initially introduced in 1813 by the French botanist J.V. Lamouroux to characterize a diverse group of eight red seaweeds within the order Floridées. Lamouroux’s classification was based on the algae’s purple to reddish coloration and the occurrence of ‘fructification’ (cystocarps) at the ends of both branches and branchlets. He chose the name in honor of his friend, a Navy official and natural scientist Mr. de La Laurencie. Although Lamouroux (1813) did not designate a type for the genus, Schmitz (1889) addressed this issue later by establishing the tribe Laurenciae and formally assigning the type species Laurencia obtusa (Hudson) Lamouroux to the genus.
For decades, the taxonomy of Laurencia has been based on its external morphology. J. Agardh (1876) first proposed subdividing the genus into four sections: Filiformes, Papillosae, Obtusae, and Pinnatifidae. Subsequently, Kylin (1923) provided a more detailed description of the reproductive structures of some species. Yamada (1931) mainly described anatomical characteristics, such as the presence of a projection of epidermal cells near the end of the branchlets, the shape and arrangement of epidermal cells in the transverse section, and the presence or absence of lenticular thickenings in medullary cells. On this basis, Yamada proposed four sections of this genus, Pinnatifidae, according to the section of J. Agardh (section with a clearly compressed frond and not palisade-like appearance of epidermal cells in the transverse section) and three new sections: Palisadae (a section of cylindrical species with palisade-like epidermal cells in the transverse section and absence of lenticular thickenings), Forsterianae (a section of cylindrical or slightly compressed species with epidermal cells that are not palisade-like and with abundant lenticular thickening), and Cartilagineae (a section of cylindrical or slightly compressed species with non-palisade-like epidermal cells without lenticular thickenings).
Over several years, this genus has undergone several nomenclatural changes (Table 1), and until recently, the Laurencia complex comprised eight genera: Laurencia sensu stricto J.V. Lamouroux, Osmundea Stackhouse, Corynecladia J. Agardh, Chondrophycus (Tokida and Saitoi) Garbary and Harper, Palisada (Yamada) K.W. Nam, Yuzurua (K.W. Nam) Martin-Lescanne, Laurenciella Cassano, Gil-Rodríguez, Senties, Diaz-Larrea, M.C. Oliveira and M.T. Fujii, and Ohelopapa F. Rousseau, Martin-Lescanne, Payri and L. LeGall.
Table 1.
The nomenclatural history of Laurencia complex
| References | Genus |
Subg enus | Section | Note |
| Lamouroux 1813 | Laurencia |
Erected genus Laurencia (including 8 species, most of which were transferred from Fucus) based on the presence of cystocarps at the end of the branches and branchlets. | ||
| Stackhouse 1816 | Pinnatifida |
Genus Pinnatifida was proposed (with Pinnatifida vulgaris as the type species) with the description: fronds gelatinous, bi-or tri-pinnate; branch obtuse; the seeds are immersed in the tips. | ||
| Schmitz 1889 | Laurencia | Designated L. obtusa Lamouroux as the type species of the genus Laurencia | ||
| Agardh 1863, Agardh 1879 | Laurencia |
Pinnatifidae; Filiformes; Papillosae; Obtusae |
Placed genus Pinnatifida Stackhouse as a section in genus Laurencia with other three sections based primarily on branching pattern and thallus compression. | |
| Yamada 1931 | Laurencia |
Pinnatifidae; Palisadae; Fosterianae; Cartilagineae |
Regroup Agardh’s sections, with section Pinnatifidae being the only remaining, and added three new sections based on the palisade formation of the cells, lenticular thickenings, and thallus compression. This was also the first study to include reproductive features. | |
| Saito 1967 | Laurencia | Laurencia |
Laurencia; Pinnatifidae; Fosterianae |
The genus is divided into subgenera, Laurencia and Chondrophycus, based on the arrangement of tetrasporangia and the presence of secondary pit connections between cortical cells in the longitudinal section. |
| Chondrophycus |
Chondrophycus; Palisadae | |||
| Saito & Womersley 1974 | Laurencia | Laurencia | Laurencia; Planae |
Introduced a new section, Planae, based on the removal of the lenticular thickenings as a significant morphological character and removed the sections of Fosterianae and Pinnatifidae. |
| Chondrophycus |
Chondrophycus; Palisadae | |||
| Nam et al. 1994 | Laurencia |
The genus Osmundea was proposed by Stackhouse in 1809 based on O. expansa (later determined to be synonymous with L. osmuda). Laurencia was conserved over Osmundea due to the popularity of the name. Then, Nam Resurrected genus Osmundea with O. osmunda as the type species. | ||
| Osmundea | The resurrection of Osmundea as a genus based on the origin of spermatangial branches and the origin of tetrasporangia. | |||
| Garbary and Harper 1998 | Laurencia | Chondrophycus was raised to a genus based on the number of pericentral cells, the presence or absence of secondary pit connections within epidermal cells, and the origin of tetrasporangia. In total, three genera (Laurencia, Chondrophycus, Osmundea) were recognized. | ||
| Osmundea | ||||
| Chondrophycus | ||||
| Nam 1999 | Laurencia |
Proposed four subgenera within Chondrophycus based on morphological characters, including the presence of secondary pit connections, the number of sterile pericentral cells in a tetrasporangial axial segment, and the arrangement of tetrasporangia. | ||
| Osmundea | ||||
| Chondrophycus | ||||
| Chondrophycus | ||||
| Kangjaewonia | ||||
| Palisada |
Palisadae, Papillosae | |||
| Yuzurua |
Parvipapillatae; Yuzurua | |||
| Nam 2006, Nam 2007 | Laurencia; Osmundea; Chondrophycus; Palisada |
Separated Palisada as a new genus. According to the previous study, subgenera Yuzurua and Palisada were merged to the new genus Palisada, and subegenera Chondrophycus and Kangjaewonii to genus Chondrophycus. | ||
| Martin-Lescanne et al. 2010 | Laurencia; Osmundea; Chondrophycus; Palisada; Yuzurua |
Resurrected and elevated subgenus Yuzurua to the generic rank, with Y. poiteaui (basionym Ch. poiteaui) designated as the type species, based on the based on the molecular analysis (rbcL). | ||
| Cassano et al. 2012c | Laurencia; Osmundea; Chondrophycus; Palisada; Yuzurua; Laurenciella |
Established the new genus Laurenciella, with La. marilzae (basionym L. marilzae) designated as the type species based on the molecular analysis (rbcL). | ||
| Metti et al. 2015 | Laurencia; Osmundea; Chondrophycus; Palisada; Yuzurua; Laurenciella; Coronaphycus |
Established the new genus Coronaphycus with Coronaphycus elatus (basionym L. elata) designated as the type species based on the molecular analysis (rbcL) and the presence of a secondary cortex in Coronaphycus. | ||
| Rousseau et al. 2017 | Laurencia; Osmundea; Chondrophycus; Palisada; Yuzurua; Laurenciella; Coronaphycus; Ohelopapa |
Establish the new genus Ohelopapa, with O. flexilis (basionym L. flexilis) as the type species based on the molecular analyses (rbcL and COI-5P) and morphological characteristics: four pericentral cells per axial cells; however, it lacks of a corps en cerise in cortical cells and secondary pit connections between cortical cells. | ||
| Cassano et al. 2019 | Laurencia; Osmundea; Chondrophycus; Palisada; Yuzurua; Laurenciella; Ohelopapa; Corynecladia |
Proposed the emendation of generic delineation of Corynecladia with C. clavata as the type species, including the newly genus Coronaphycus based on the molecular analysis (rbcL), specimens identified as L. clavata (which was previously belonged to Corynecladia, a genus proposed by Agardh in 1876) were grouped with seqeunces of Coronaphycus elatus and Coronaphycus novus. Due to priority over Coronaphycus, Corynecladia was selected as the valid genus, encompassing three species: C. clavata, C. elata, and C, nova. |
Osmundea was originally proposed by Stackhouse (1809) but was recognized and resurrected as a genus within Laurencia complex decades later by Nam et al. (1994) based on the number of pericentral cells per axial cell (two, rather than four in Laurencia s.s.), the origin of tetrasporangia, the origin and branching of the spermatangial branch, the shape of apical spermatangial pits, and the alignment of presporangial cover cells. Garbary and Harper (1998) elevated the subgenus Chondrophycus as a sister taxon to Osmundea because of two pericentral cells per axial cell and different from Laurencia s.s. by the absence of secondary pit connections between adjacent cortical cells. A corps en cerise was also diagnosed in the study of Chondrophycus, however, because this structure has only been observed in living materials, this characteristic was not useful for generic diagnostics at the time. Based on Yamada’s (1931) section Palisadae, many species that were previously nested within Chondrophycus have been transferred to Palisada, a genus proposed and validated by Nam (2007), with tetrasporangial development being unique to Chondrophycus. The five characteristics that separate Palisada from other genera in the Laurencia complex are the first pericentral cell position compared to the trichoblast, the second pericentral cell’s fertility, spermatangial branch patterns during formation on trichoblasts, auxiliary cell timing, and the number of pericentral cells of the procarp-bearing segment (Nam 2006).
The four genera accepted in the Laurencia complex as mentioned above were all established based on morphological characteristics, while early molecular research also recognized other genera within the complex. Yuzurua segregated from Palisada and was the first genus within the complex to be identified based on molecular evidence (rbcL). It is morphologically different from Palisada because of the absence of palisade-like cells and the lack of secondary pit connections in cortical cells. The second genus to be identified by molecular evidence was Laurenciella (rbcL) (Cassano et al. 2012b). This genus is morphologically cryptic compared to its sister genus, Laurencia s.s. Metti et al. (2015) established the genus Coronaphycus using molecular (rbcL) and morphological features, but it was later determined to be conspecific with Cornynecladia J. Agardh 1876, which had priority (Cassano et al. 2019). The last genus to be established was Ohelopapa, which was recognized based on both molecular analysis (rbcL and COI-5P) and morphological characteristics, i.e., four pericentral cells per axial cell, however, it lacks a corps en cerise in cortical cells and secondary pit connections between cortical cells (Rousseau et al. 2017). The corps en cerise is a refraction-specific intracellular organelle that is used as the main storage site for halogenated compounds in the Laurencia complex (Fujii et al. 2012). The corps en cerise has been found in Laurencia s.s. (Masuda et al. 1996, Cassano et al. 2012a, Francis et al. 2017, Metti 2017, Metti 2022) and Laurenciella (Gil-Rodríguez et al. 2009 and Rocha-Jorge et al. 2010 as Laurencia marilzae Gil-Rodríguez, Senties et M.T. Fujii; Collado-Vides et al. 2018), while other genera within the complex do not possess this structure. It has recently been used as a generic characteristic for identification within the Laurencia complex.
Taxonomy of the Genus Laurencia
The taxonomy of the genus has undergone significant revision, with emphasis placed on characteristics such as color, presence or absence of secondary pit connections between cortical cells, arrangement of tetrasporangia, presence or absence of lenticular thickenings, projection of cortical cells, and presence of a corps en cerise.
Color is considered a characteristic for identifying species within the genus Laurencia. In a study by Gil-Rodríguez and Haroun (1992), Laurencia viridis Gil-Rodríguez and Haroun was compared with other Laurencia species from the Canaries (i.e., L. obtusa, L. majuscula, L. minuta, and L. tenera) among which only L. viridis has a bright green color. It was then compared with other green species of Laurencia from other parts of the world: i.e., L. intricata, L. flexilis, L. nidifica, L. “green”, L. okamurai, and L. intermedia. Although Garbary and Harper (1998) did not include color as a diagnostic characteristic of Laurencia, they proposed that it is a useful characteristic when color differences are examined with regard to particular species complexes.
Saito (1967) highlighted the presence of a secondary pit-connection among superficial cortical cells (Fig. 1A) as a distinctive feature for identifying the Laurencia complex; however, this feature was absent in Chondrophycus. Another key criterion for species differentiation within Laurencia is the presence or absence of lenticular thickening on the medullary cells (Fig. 1B). This characteristic has been recognized as species-specific, as illustrated by the occurrence of lenticular thickening in Laurencia forsteri (Mertens ex Turner) Greville (Saito and Womersley 1974). Garbary and Harper (1998) suggested the inclusion of these characteristics in morphological studies of genera and species within the Laurencia complex. The presence or absence of projecting cells at branch apices (Fig. 1C) is another valuable characteristic of Laurencia. Several species can be identified by conspicuously projecting superficial cells, through which they can be differentiated from related species, including Laurencia moretonensis A.B. Cribb which was distinguished from Laurencia pannosa Zanardini by having no projecting cells near the apical cells (Cribb 1958), Laurencia pinnata Yamada, which differs from Laurencia mariannensis Yamada by the presence of projecting cells (Yamada 1931), and Laurencia saitoi Perestenko, which differs from L. obtusa also by having projecting cells (Masuda & Abe 1993).
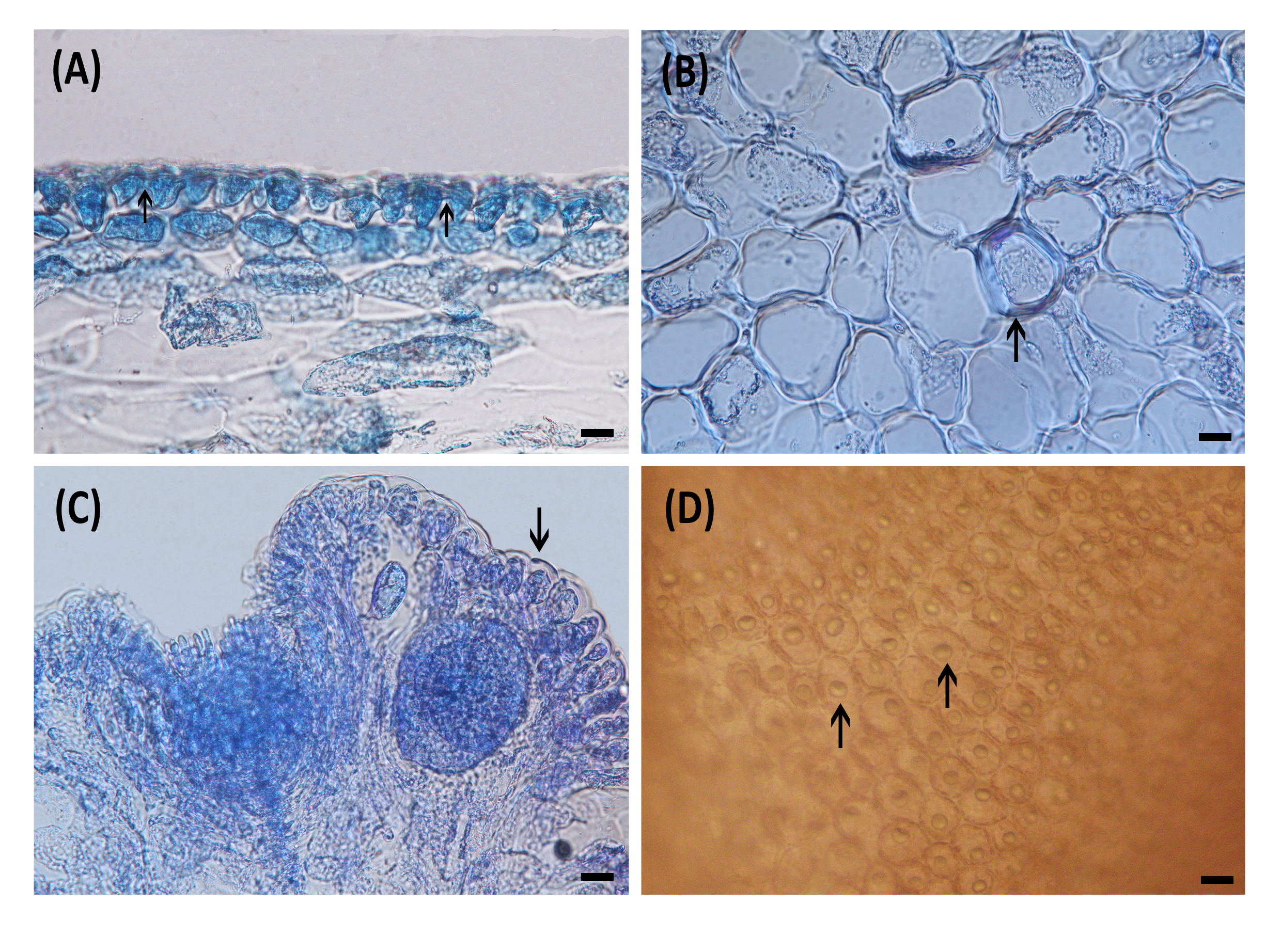
Fig. 1.
Characteristics of Laurencia. (A) Secondary pit connections between cortical cells in longitudinal section (arrows), (B) Presence of lenticular thickenings on medullary cells (arrow), (C) Projecting cells near the apical cells in longitudinal section (arrow), (D) Presence of single corps en cerise per medullary cells in surface view (arrows). Scale bars: (A-D) 20 mm.
Laurencia s.s. stands out by having distinct intracellular organelles known as “corps en cerise” (Fig. 1D), characterized by their unique refractive properties, however, the genus Laurenciella shares some morphological features with Laurencia, including the presence of a corps en cerise. The crucial point of differentiation between the two genera is the presence of a corps en cerise in all cells (cortical, medullary, pericentral, axial, and trichoblast cells) in Laurenciella marilzae (Gil-Rodríguez, Senties, Diaz-Larrea, Cassano & M.T. Fujii) Gil-Rodríguez, Senties, Diaz-Larrea, Cassano & M.T. Fujii (Gil-Rodríguez et al. 2009). The presence or absence and number of corps en cerises per cell are valuable diagnostic characteristics at the species level (Masuda 1997), as observed in Laurencia brongniartii J. Agardh, which is distinguished from L. pinnata by the presence of two or three corps en cerises per cell, in contrast to the single corps en cerise in L. pinnata (Abe et al. 1998). Despite its importance in characterizing Laurencia species, this feature is only evident in very young plants (Masuda et al. 1997) and remains undetected in liquid-preserved and dried specimens, limiting such observations to living specimens (McDermid 1988).
Taxonomic studies of the Laurencia complex have traditionally relied on morphological characteristics such as branching patterns, tetrasporangial structures, and reproductive organs, however, this approach has some limitations with regard to morphological plasticity and convergent evolution within the genus. Laurencia has been reported to produce more than 250 diverse compounds (Abe and Masuda 1998) and related or unrelated sets of metabolites in different populations. Based on many morphologically similar but chemically distinct taxa, chemotaxonomy has been used as a tool to identify Laurencia (Abe et al. 1975, Fenical and Norris 1975, Masuda et al. 1996, Masuda et al. 2002).
Diversity and Distribution in East Asia and Southeast Asia regions
The genus Laurencia currently includes 144 taxonomically accepted species found globally in temperate to tropical waters, and inhabiting littoral to sublittoral areas (Guiry and Guiry 2023). Most of the species within this genus have been reported as type localities in Australia (13 species), South Africa (13 species), Mexico (11 species), Japan (8 species), and California (7 species). In accordance with McDermid (1988), the distribution of Laurencia species is concentrated in the Southern Hemisphere (Fig. 2).
Checklists were compiled for the Asia North-Pacific region: Korea, Japan, and China in East Asia; and Indonesia, Malaysia, Myanmar, Philippines, Thailand, and Vietnam in Southeast Asia (Table 2). A total of 47 Laurencia species were reported in the East Asia and Southeast Asia regiona, with 14 species recorded in Korea (Nam 2011), 18 species in Japan (Yoshida et al. 2015), 20 species in China (Liu 2008), 17 species in Indonesia (Atmadja and van Reine 2012), 9 species in Malaysia (Phang et al. 2016), 4 species in Myanmar (Soe-Htun et al. 2021), 22 species in Philippines (Lastimoso and Santiañez 2021), 1 species in Thailand (Tsutsui et al. 2012), and 22 species in Vietnam (Nguyen et al. 2023) (Fig. 3).
Table 2.
Checklist of Laurencia from the Asia North-Pacific with support of morphological and molecular observations
| Taxa No | Type locality | Korea1 | Japan2 | China3 | Indonesia4 | Malaysia5 | Myanmar6 | Philippines7 | Thailand7 | Vietnam9 | DNA data | References | |
| 1 | Laurencia botryoides (C.Agardh) Gaillon | Australia | na | na | |||||||||
| 2 | Laurencia brachyclados Pilger | West Africa | + |
23S rRNA, 28S rRNA, COI | Sherwood et al. 2010 | ||||||||
| 3 | Laurencia brongniartii J.Agardh | Martinique | + | + | + | + | rbcL | Fujii et al. 2006, Gil-Rodríguez et al. 2010, Metti et al. 2015 | |||||
| 4 | Laurencia caduciramulosa Masuda & S.Kawaguchi | Vietnam | + | + | + | rbcL | Cassano et al. 2012c, Collado-Vides et al. 2014 | ||||||
| 5 | Laurencia calliclada Masuda | Vietnam | + | + | na | na | |||||||
| 6 | Laurencia caraibica P.C.Silva | Bahamas | + | rbcL | Gil-Rodríguez et al. 2009 | ||||||||
| 7 | Laurencia chinensis C.K.Tseng | China | + | + | cox1 | Unpublished | |||||||
| 8 | Laurencia chondrioides Børgesen | Caribbean, USA | + | na | na | ||||||||
| 9 | Laurencia composita Yamada | Japan | + | + | + | + | + | na | na | ||||
| 10 | Laurencia corymbosa J.Agardh | South Africa | + | rbcL | Francis et al. 2017* | ||||||||
| 11 | Laurencia decumbens Kützing | New Caledonia | + | + | + | + | + |
23S rRNA, 28S rRNA | Sherwood et al. 2010 | ||||
| 12 | Laurencia dendroidea J.Agardh | Brazil | + | + | + | + | + | + | + | rbcL | Fujii et al. 2006*, Cassano et al. 2012c*, Machín-Sánchez et al. 2014, Popolizio et al. 2022 | ||
| COI | Machín-Sánchez et al. 2014, Popolizio et al. 2022 | ||||||||||||
| ITS | Popolizio et al. 2022 | ||||||||||||
| 13 | Laurencia distichophylla J.Agardh | New Zealand | + | na | na | ||||||||
| 14 | Laurencia filiformis (C.Agardh) Montagne | Australia | + | + | + | + | SSU | Phillips et al. 2000 | |||||
| rbcL | Cassano et al. 2019 | ||||||||||||
| 15 | Laurencia forsteri (Mertens ex Turner) Greville | New Zealand | + | na | na | ||||||||
| 16 | Laurencia galtsoffii M.Howe | Hawaii | + | + |
23S rRNA, 28S rRNA, COI | Sherwood et al. 2010* | |||||||
| 17 | Laurencia glandulifera (Kützing) Kützing | Italy | + | + | + | + | na | na | |||||
| 18 | Laurencia glomerata (Kützing) Kützing | South Africa | + | rbcL | Díaz‐Tapia et al. 2017, Francis et al. 2017*, Mshiywa et al. 2023 | ||||||||
| 19 | Laurencia hamata Yamada | Japan | + | + | na | na | |||||||
| 20 | Laurencia heteroclada Harvey | Australia | + | rbcL, COI | Metti 2022* | ||||||||
| 21 | Laurencia hongkongensis C.K.Tseng, Chang, E.Z.Xia & B.M.Xia | Hong Kong | + | na | na | ||||||||
| 22 | Laurencia intercalaris K.W.Nam | Korea | + | na | na | ||||||||
| 23 | Laurencia intricata J.V.Lamouroux | Antilles | + | + | + | + | + | + | rbcL | Fujii et al. 2006, Cassano et al. 2009, Gil-Rodríguez et al. 2009, Cassano et al. 2012a*, Collado-Vides et al. 2018, Popolizio et al. 2022 | |||
| COI | Popolizio et al. 2022 | ||||||||||||
| 24 | Laurencia japonensis T.Abe & Masuda | Japan | + | na | na | ||||||||
| 25 | Laurencia lageniformis Masuda & Suzuki | Vietnam | + | + | na | na | |||||||
| 26 | Laurencia laxa (R.Brown ex Turner) Gaillon | South Africa | + | na | na | ||||||||
| 27 | Laurencia mariannensis Yamada | Saipan | + | + | + | + | + | rbcL | Martin-Lescanne et al. 2010 | ||||
| 28 | Laurencia microcladia Kützing | West Indies | + | rbcL, COI, ITS | Popolizio et al. 2022 | ||||||||
| 29 | Laurencia nangii Masuda | Vietnam | + | na | na | ||||||||
| 30 | Laurencia natalensis Kylin | South Africa | + | rbcL | Fujii et al. 2006, Francis et al. 2017*, Garcia-Soto 2017, Kundu 2022 | ||||||||
| COI | Garcia-Soto 2017, Kundu 2022 | ||||||||||||
| 31 | Laurencia nidifica J.Agardh | Hawaii | + | + | + | + | + | + | SSU, LSU, cox1 | Kurihara et al. 2010* | |||
|
23S rRNA, 28S rRNA, COI | Sherwood et al. 2010* | ||||||||||||
| 32 | Laurencia nipponica Yamada | Japan | + | + | + | + | SSU, LSU, cox1 | Kurihara et al. 2010* | |||||
| 33 | Laurencia obtusa (Hudson) J.V.Lamouroux | England, UK | + | + | + | + | + | + | + | + | rbcL | Nam et al. 2000, Fujii et al. 2006, Rousseau et al. 2017, Preuss et al. 2023 | |
| COI | Rousseau et al. 2017 | ||||||||||||
| 34 | Laurencia okamurae Yamada | Japan | + | + | + | + | na | na | |||||
| 35 | Laurencia omaezakiana Masuda | Japan | + | + | na | na | |||||||
| 36 | Laurencia pannosa Zanardini | Malaysia | + | + | na | na | |||||||
| 37 | Laurencia pinnata Yamada | Japan | + | + | + | + | + | + | + | + | na | na | |
| 38 | Laurencia saitoi Perestenko | Russia | + | + | COI | Saunders 2014 | |||||||
| 39 | Laurencia silvae J.F.Zhang & B.M.Xia | China | + | + | na | na | |||||||
| 40 | Laurencia similis K.W.Nam & Y.Saito | Australia | + | + | na | na | |||||||
| 41 | Laurencia snackeyi (Weber Bosse) M.Masuda | Indonesia | + | + | + | 18S rRNA, cox1 | Díaz‐Tapia et al. 2017 | ||||||
| 42 | Laurencia subsimplex C.K.Tseng | Hong Kong | + | + | na | na | |||||||
| 43 | Laurencia succulenta K.W.Nam | Korea | + | na | na | ||||||||
| 44 | Laurencia tenera C.K.Tseng | Hong Kong | + | + | 28S rRNA | Sherwood et al. 2010 | |||||||
| 45 | Laurencia tristicha C.K.Tseng, C.F.Chang, E.Z.Xia & B.M.Xia | Hong Kong | + | na | na | ||||||||
| 46 | Laurencia tropica Yamada | Saipan | + | + | + | + | + | na | na | ||||
| 47 | Laurencia venusta Yamada | Japan | + | + | + | rbcL | Díaz-Larrea et al. 2007, Metti et al. 2015 |
References: 1Nam 2011, 2Yoshida et al. 2015, 3Liu 2008, 4Atmadja & van Reine 2012, 5Phang et al. 2016, 6Soe-Htun et al. 2021, 7Lastimoso & Santiañez 2021, 8Tsutsui 2012, 9Nguyen et al. 2023.
Fig. 3.
The maximum likelihood (ML) phylogenetic tree based on rbcL gene of Laurencia (Popolizio et al. 2022).
Both L. obtusa (type locality: England) and L. pinnata (type locality: Japan) are widely distributed, covering most areas except Singapore and Thailand. The distribution of marine species can also be driven by ocean currents (Li et al. 2017), and the wide distribution of Laurencia in Asia North-Pacific waters likely influenced by the North-South-Equatorial Current that runs along the Pacific Ocean and the Kuroshio Current, which could influence long-distal dispersal of multiple marine species from south to north (Yamasaki et al. 2014).
In contrast, Laurencia hongkonensis C.K. Tseng, Chang, E.Z. Xia & B.M. Mia (type locality: Hong Kong) and Laurencia tristicha C.K. Tseng, C.F. Chang, E.Z. Xia & B.M. Mia (type locality: Hong Kong) exhibited endemism, and were found exclusively in their respective localities. This endemism was further supported by Tseng (1983), who found that these two species were reported only in China (Hong Kong). Additionally, Laurencia intercalaris Nam (type locality: Korea) and Laurencia succulenta Nam (type locality: Korea) are restricted to Korea, whereas Laurencia japonensis T. Abe & Masuda (type locality: Japan) are endemic to Japan.
Molecular Phylogenetic Research
The notable morphological plasticity of individuals, the challenge of observing specific vegetative or reproductive features, and the presence of inconsistent and incomplete species descriptions have contributed to the considerable ambiguity in the identification, classification, and nomenclature of organisms allegedly comprising this complex. Recent DNA barcoding and phylogenetic analyses have enabled researchers to reconstruct the evolutionary history of Laurencia with remarkable accuracy. Over the past two decades, the taxonomy of the Laurencia complex has markedly advanced through integrated application of anatomical and molecular markers on a global scale.
Molecular studies on the Laurencia complex were cconducted using various genetic markers, including SSU (Phillips et al. 2000, Kurihara et al. 2010, Díaz-Tapia et al. 2017, Ortega et al. 2020), LSU (Kurihara et al. 2010, Sherwood et al. 2010, Du et al. 2015, Rousseau et al. 2017), UPA (Sherwood et al. 2010, Du et al. 2015), and ITS (Lewis et al. 2008, Popolizio et al. 2022). Although several mmarkers have been used in these studies, molecular analyses of the Laurencia complex have predominantly relied on the plastid marker rbcL and the mitochondrial marker COI. Molecular phylogenetic studies have led to the identification and reinstatement of a cconsiderable number of new or previously misidentified species (Cassano et al. 2012a, Metti et al. 2015, Francis et al. 2017, Cassano et al. 2019, Popolizio et al. 2022).
Within the Laurencia complex, Yuzurua, Laurenciella, Ohelopapa, and Corynecladia have been identified using molecular analyses (Martin-Lescanne et al. 2010, Cassano et al. 2012c, Rousseau et al. 2017, Cassano et al. 2019). The identifiaction of these genera relied on rbcL gene, in additional to the COI-5P gene, specifically with regard to Ohelopapa. These molecular insights have contributed to a better understanding of the diversity within the Laurencia complex, highlighting the importance of molecular markers for unraveling the evolutionary relationships and taxonomy of these algae.
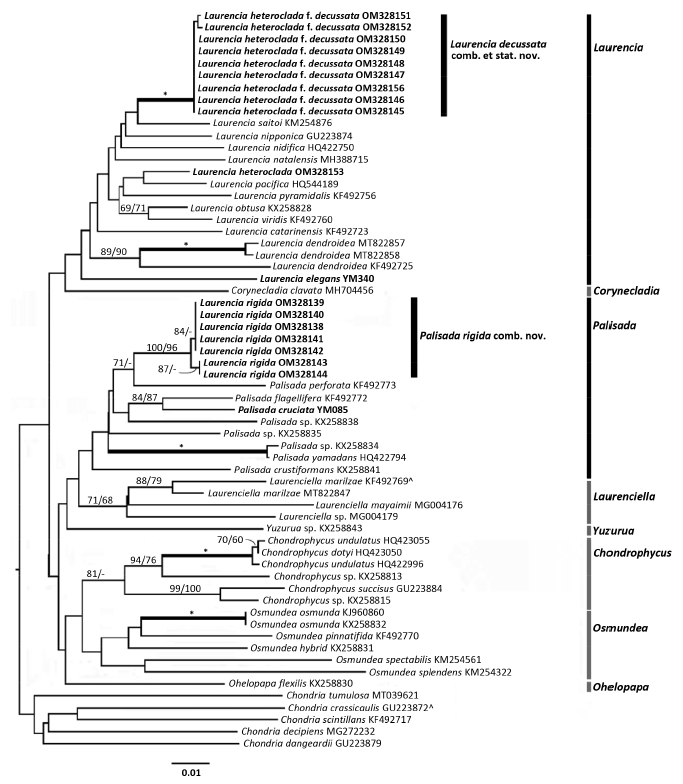
Through molecular analyses, Popolizio et al. (2022) proposed two new species, Chondrophycus planiparvus Popolizio, C.W. Schneider & C.E. Lane and Laurenciella namii Popolizio, C.W. Schneider & C.E. Lane (Fig. 4). Remarkably, L. namii shows genetic differences from other species in the genus Laurenciella. Additionally, the study supported the recognition of two species, L. dendroidea and L. catarinensis Cordeiro-Marino & M.T. Fujii, which has historically been identified as L. obtusa in Bermuda.
Molecular studies based on rbcL gene have revealed that Laurencia majuscula (Harvey) A.H.S.Lucas from Australia is conspecific to Laurencia dendroidea J. Agardh from Brazil, despite their disjunct-type localities, in which L. dendroidea has priority (Metti et al. 2013). Following a study on L. majuscula, its non-typical variety L. majuscula var. elegans (A.H.S.Lucas) Saito & Womersley, formed a separate, well-supported clade from L. dendroidea based on COI-5P and rbcL sequences. It has since been reinstated at the species level as Laurencia elegans A.H.S. Lucas (Metti 2017). Recently, Metti (2022) reported a significant molecular distance between L. heteroclada f. decussata and L. heteroclada, supporting the separation of L. heteroclada f. decussata into a new species, Laurencia decussata (Fig. 5).
Until recently, most molecular studies on Laurencia were conducted in the Atlantic Ocean, Northeast Pacific, and Southwest Pacific, whereas very limited research has been conducted in the Asia Pacific (Table 3). Several Laurencia species reported have type localities in the Asia-Pacific region, including Japan, Hong Kong, Korea, and Vietnam, however, studies on this taxon are primarily confined to morphological analyses, with limited phylogenetic studies to date. Kurihara et al (2010) conducted molecular phylogenetic studies focused on red algal parasites, including Janczewskia, parasitic red algae of Laurencia collected from the Hawaiian Islands, Japan, and Russia. Since then, there have been no updated molecular phylogenetic studies of Laurencia in the Asia-Pacific region.
Table 3.
Molecular studies of Laurencia complex conducted to date
| Taxa | Markers | Region | References |
| L. glomerata | rbcL | South Africa | Mshiywa et al. 2023 |
| L. tasmanica, L. obtusa | rbcL | Australia; Spain | Preuss et al. 2023 |
| L. obtusa, L. catarinensis | cox1 | Spain | |
| L. microcladia, L. intricata, L. dendroidea, L. catarinensis | rbcL | Bermuda; USA | Popolizio et al. 2022 |
| L. microcladia, L. intricata,, L. dendroidea, L. catarinensis | COI | ||
| L. microcladia, L. dendroidea, L. catarinensis | ITS | ||
| L. decussata*, L. heteroclada | rbcL | Australia | Metti 2022 |
| L. decussata, L. heteroclada, L. elegans | COI | ||
| L. flexuosa | rbcL | India | Kundu 2022 |
| L. natalensis, L. flexuosa | COI | ||
| L. dendroidea, La. marilzae | COI; rbcL | Australia; Italy | Serio et al. 2020 |
| L. obtusa | SSU | Saudi Arabia | Ortega et al. 2020 |
| L. digitata, Ch. anabeliae | rbcL | Venezuela | Cassano et al. 2020 |
| L. mutuae* | rbcL; COI | Mexico | Sentíes et al. 2019 |
| L. filiformis, L. longiramea*, Corynecladia clavata^ (as L. clavata) | rbcL | Australia; Brazil | Cassano et al. 2019 |
| L. karachiana* | rbcL; COI | Pakistan | Bibi et al. 2019 |
| L. intricata, Laurenciella mayaimii^, Laurenciella sp. | rbcL | USA | Collado-Vides et al. 2018 |
| L. obtusa, L. dendroidea (as L. cf. majuscula), Laurencia sp. (as L. cf. brongniartii, L. cf. calliptera, L. cf. mariannensis, L. cf. nidifica), Laurencia sp. (as L. cf. natalensis), Laurencia sp. (as L. cf. flexuosa), La. marilzae, Ohelopapa flexilis^, O. oederi, O. hybrida, O. osmunda, Palisada sp. (as P. cf. parvipapillata, P. cf. robusta), P. crustiformans* | rbcL; COI; LSU |
Croatia; France; New Caledonia; Oman; South Africa, Sri Lanka; Tahiti; USA | Rousseau et al. 2017 |
| L. natalensis, L. dendroidea, L. caraibica | rbcL | Venezuela | Garcia-Soto 2017 |
| L. natalensis, L. dendroidea | COI | Venezuela | |
| L. dichotoma*, L. alfredensis*, L. stegengae*, L. sodwaniensis*, L. pumila var. dehoopiensis*, L. digitata*, L. multiclavata*, L. natalensis, L. pumila, L. glomerata, L. flexuosa, L. complanata, L. corymbosa, L. cf. brongniartii, Chondrophycus sp., Palisada sp., La. marilzae | rbcL | South Africa | Francis et al. 2017 |
| L. snackeyi, Palisada sp. | 18S rRNA | Australia | Díaz-Tapia et al. 2017 |
| L. glomerata, L. tasmanica, Laurenciella sp., Corynecladia clavata (as L. clavata) | rbcL | Australia; South Africa | |
| L. snackeyi, Palisada sp. | cox1 | Australia | |
| L. venusta, L. brongniartii, Chondrophycus sp., Corynecladia elata (as Coronaphycus elatus^), Corynecladia nova (as Ccoronaphycus novus) | rbcL | Australia | Metti et al. 2015 |
| L. obtusa, L. nipponica, C. intermedius |
UPA/23S rRNA; LSU | China | Du et al. 2015 |
| L. pacifica, L. saitoi | COI | USA | Saunders 2014 |
| L. pyramidalis, L. viridis, L. dendroidea, L. catarinensis, La. marilzae | rbcL |
Spain; France; Portugal | Machín-Sánchez et al. 2014 |
| L. pyramidalis, L. viridis, L. dendroidea, L. catarinensis, O. pinnatifida, P. flagellifera, P. perforata, La. marilzae | COI |
Spain; France; Portugal | |
| L. caduciramulosa | rbcL | USA | Collado-Vides et al. 2014 |
| L. aldingensis, L. caduciramulosa, O. truncata | rbcL | Brazil; Spain | Cassano et al. 2012c |
| L. oliveirana | rbcL | Brazil | Cassano et al. 2012b |
| L. dendroidea, L. intricata, P. flagellifera, P. furcata | rbcL | Brazil; Spain; Cuba | Cassano et al. 2012a |
| L. mcdermidiae, L. nidifica, L. galtsoffii, L. brachyclados, L. dendroidea (as L. majuscula), Laurencia sp., P. yamadana, P. cartilaginea, P. parvipapillata, P. crustiformans (as L. "crustiformans), C. cf. undulatus, Chondrophycus sp. |
UPA/23S rRNA; LSU; COI | Hawaii | Sherwood et al. 2010 |
| L. caduciramulosa | rbcL | Cuba | Sentíes et al. 2010 |
| L. pyramidalis, L. mariannensis, O. hybrida, O. osmunda | rbcL |
UK; New Caledonia; France | Martin-Lescanne et al. 2010 |
| L. brongniartii, L. intricata, P. flagellifera | rbcL |
Australia; Mexico; Spain | Gil-Rodríguez et al. 2010 |
| L. mcdermidiae, L. nidifica, L. nipponica, L. dendroidea (as L. majuscula), Laurencia sp. 1, Laurencia sp. 2, Laurencia sp. 3, Laurencia sp. 4, Laurencia sp. 5, Laurencia sp. 6, Laurencia sp. 7 |
SSU; LSU; cox1 | USA; Japan; Russia | Kurihara et al. 2010 |
| Laurenciella marilzae (as L. marilzae)*, L. caraibica, L. intricata, L. viridis, L. dendroidea (as L. majuscula), Laurencia sp., Chondrophycus sp., O. pinnatifida, P. perforata (as L. papillosa) | rbcL | Mexico; Spain | Gil-Rodríguez et al. 2009 |
| P. perforata, L. intricata | rbcL |
Brazil; Mexico; Spain | Cassano et al. 2009 |
| L. viridis |
ITS1; ITS2; rbcL | Spain | Lewis et al. 2008 |
| L. venusta | rbcL | Mexico | Díaz-Larrea et al. 2007 |
| L. pacifica, L. natalensis, L. filiformis, L. flexuosa, L. complanata, L. brongniartii, L. intricata, L. obtusa, L. dendroidea, L. translucida | rbcL |
Brazil; Mexico; Guadeloupe; USA; Venezuela; South Africa; Taiwan | Fujii et al. 2006 |
| L. filiformis | 18S rRNA | Australia | Phillips et al. 2000 |
| L. obtusa | rbcL | Ireland | Nam et al. 2000 |
An initial study of the Laurencia complex based on a taxon-rich dataset (rbcL) and a character-rich dataset (COI-5P + rbcL + LSU) revealed diverse taxonomy at the genus level within the Laurencia complex, which also led to the establishment of the Ohelopapa genus, previously identified as Laurencia (Rousseau et al. 2017). This confirms that molecular-assisted studies are an effective approach for precisely determining the taxonomic positions of some members of the Laurencia complex. Recent studies on the plastid genome of Laurencia by Verbruggen and Costa (2015) (L. snackeyi) and Preuss et al. (2023) (L. obtusa, L. catarinenis, Co. elata, and some new species identified as Janckzewskia) have marked significant advancements and hold the potential to address the challenges associated with unresolved phylogenetic relationships within the genus Laurencia.
Life History
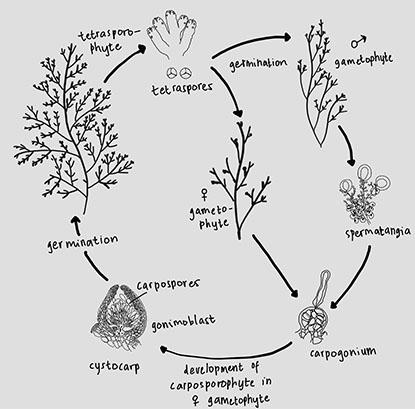
The life cycles of Florideophiceae can be heteromorphic, meaning that they differ from one another, or isomorphic, meaning that they are identical (Hawkes 1990); however, most red algae exhibit the same triphasic pattern (Yoon et al. 2010). The genus Laurencia s.s. reproduces sexually and has typical triphasic isomorphic life histories involving haploid sexual gametophytes, diploid carposporophytes that develop on female gametophytes, and free-living diploid tetrasporophytes that have similar or identical form phases (Bleckwenn et al. 2003).
During the life cycle of Laurencia s.s. (Fig. 5), the spermatangial stichidia release male gametes, or spermatids. Subsequently, male gametes or spermatia fuse with female gametes or carpogonia. Carpospores are released after male gametes or spermatia fuse with female gametes or carpogonia. Upon germination, carpospores develop into tetrasporophytes. The tetraspores are then released and germinate, giving rise to both male and female gametophytes.
Ecological Role and Utilization
Laurencia plays an ecological role in the ecosystem, serving as a habitat and refuge for other marine organisms, such as being a host for parasitic algae including Erythrocystis and Janczewskia (Kurihara et al. 2010). Laurencia is also a habitat for the young juvenile spiny lobster Panulirus argus, where juveniles use the algal microhabitat by successive settlement before moving to the substrate for attachment (Marx and Herrnkind 1985). Crabs of Microphrys bicornutus not only use L. papillosa as food but also as camouflage (Kilar and Lou 1986), while the conch species Strombus gigas feeds on Laurencia, utilizing it not only as a food source but also for the supply of chemical cues that trigger the metamorphosis of its larvae (Boettcher and Targett 1996). Among the invertebrates that feed on Laurencia, sea hares belonging to the genus Aplysia are the most common grazers. These sea hares have acquired chemical defense mechanisms from the algae, frequently in such form that they can be employed for their protection (Palaniveloo and Vairappan 2014). The green turtle (Chelonia midas) is a marine herbivore that has been reported to consume seagrass and algae, including Laurencia spp., in the Torres Strait, Australia (Garnett et al. 1985, André et al. 2005).
Laurencia has been known among local communities for centuries, with various edible species, including L. undulata (now Chondrophycus undulatus) and L. nidifica (referred to as “mane’one’o” in Hawaii) (Abbott and Williamson, 1974, Li et al. 2009) being consumed across the Pacific Ocean. In Japan, there are reports of fisherman harvesting L. nipponica and consuming it as soup after drying; however, some Japanese believe that consuming Laurencia may have abortive effects (Saito 1967).
This genus has an extraordinary diversity of structurally unique metabolites that have been consistently isolated from its representatives over the past few decades. These metabolites span a range of compound classes including sesquiterpenes, diterpenes, triterpenes, acetogenins, indoles, aromatic compounds, steroids, and miscellaneous compounds. Numerous metabolites derived from Laurencia have been assessed for their effectiveness against diverse bacterial and fungal strains and have demonstrated varying degrees of activity. These metabolites, which are sourced from different origins, undergo antimicrobial activity evaluation using various methods and / or different strains of microorganisms. Notably, isoobtusol, isoobtusol acetate, and obtusol have been identified as common constituents of several Laurencia species, and their antibacterial and antifungal activities have been documented in several studies (Suenaga 2004, Vairappan 2003). Nuclear Magnetic Resonance (NMR) spectroscopy of the other active fractions obtained from initial chromatographic separation of the extract revealed the presence of typical Laurencia compounds. This suggests that additional metabolites within these fractions may also exhibit antimalarial activity (Wright et al. 1996). Bromophenol 985 was obtained from an acetone extract of L. nipponica and demonstrated mild inhibition of glucose 6-phosphate dehydrogenase which has been explored as a potential therapeutic agent for treating obesity (Mikami et al. 2013).